Planar Polarity
|
||||
|
A central problem in cell and developmental biology is understanding how cells and groups of cells become organized into higher order structures. One form of higher organization that is currently under intense investigation is planar cell polarity. Planar cell polarity is the coordinate organization of cells within the plane of a single layered sheet of cells (see Fig 1). While cell polarity requires cells to distinguish their apical and basal-lateral surfaces, using cell-cell and cell substratum contact cues, Planar Polarity is the organization of groups of cells in the plane of the epithelium, such that they all orient to similar, apparently remote co-ordinates. Planar polarity is essential for tissue functioning: the planar polarity of the vertebrate inner ear is essential to proper hearing and balance; similarly, planar polarity of the fly eye is essential for accurate vision. We are using the fruit fly, Drosophila melanogaster, as a genetically tractable organism to investigate the genetic and molecular mechanisms underlying planar polarity.
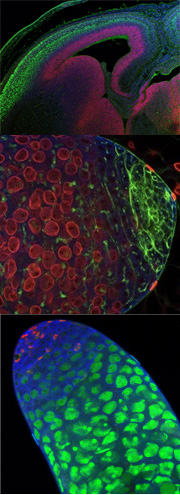
Figure 1. Planar Cell Polarity in the ear. On the left is a scanning EM of the Planar polarity is evident in Drosophila in the organization of bristles on the back, photoreceptors in the eye and hairs on the wing. (see Figure 2)
Figure 2. Examples of Planar polarized tissues in Drosophila. In the eye, each ommatidium is formed by 8 photoreceptors, R1-R8. These photoreceptors resemble an arrowhead in sections, with all the ommatida in the dorsal half of the eye pointing dorsally (see Figure 2 top panel). On the back of the fly, all of the chemosensory bristles point posteriorly (see Figure 2, bottom left panel) In the fly wing, planar polarity is evident that all of the cells secrete a single hair that points distally (Figure 2, bottom right panel). A genetic pathway responsible for the accurate organization of cells in all of these tissues has been determined through analysis of mutants that specifically affect planar polarity, while leaving cell identity.
|
||||
Mirror and Fat projects
|
||||
|
|
Dorsal-ventral Planar polarity in the fly eye
D/V planar polarity, seen in cross-sections (above right) is formed when ommatida in the eye disc rotate 90° through space , to achieve opposite polarities in the dorsal and ventral fields of the eye. How these rotations occur is still not clear.
Ommatidia rotate to achieve D/V planar polarity
Mirror Regulates D/V planar polarityWe identified a novel transcription factor, which we called Mirror, which is expressed only in the dorsal half of the eye, and regulates the precise patterning that occurs at the equatorial border of dorsal-ventral planar polarity.
Mirror expression is indicated in by the pigment granules in the sections ( left) and by the red shading ( schema on right) Where borders of Mirror expressing cells meet non-expressing cells, borders of planar polarity form. This is true for the normal equator (top) or for ectopic borders formed by clones (bottom).
Mirror regulates cell adhesionOur first hint that mirror may control cell adhesion came when we over expressed mirror in a patch of cells in the eye - furrows formed at the border between mirror expressing and non-expressing cells. Another indication that mirror was affecting cell adhesion came when we looked at rare mirror mutant flies that survived to adulthood - they displayed a bizarre ball on a ball eye, which suggested that some sort of cell-sorting was occurring, a condition that occurs when you experimentally manipulate cell-cell adhesion molecules. Another indication that mirror is controlling cell-cell interactions came when we analyzed the shape of the clones. Most clones in the eye show random, irregular borders, suggesting that cells within the clones and outside of it mix randomly. If we looked at fringe mutant clones in the ventral half of the eye, we saw that these clones, too, mix with surrounding cells, suggesting that in the eye, fringe is not controlling cell-cell interactions. However when we examined the shapes of mirror mutant clones, we saw that they have very regular, smooth borders. This indicates that cells expressing mirror do not mix with surrounding cells: they appear to have different affinities. Consistent with this being a fringe-independent function of mirror, double mutant clones show the same phenotype as mirror mutant clones. |
|||
Publications:
|
||||
|
|
||||
 |