

HOME
RESEARCH
PUBLICATIONS
PEOPLE
CONTACT INFO
ProteoSync
ProteoSync
ProteoSync is a sequence alignment tool created by Elliot Sicheri that allows comparison and visualization of protein sequence conservation across a wide range of selectable species.
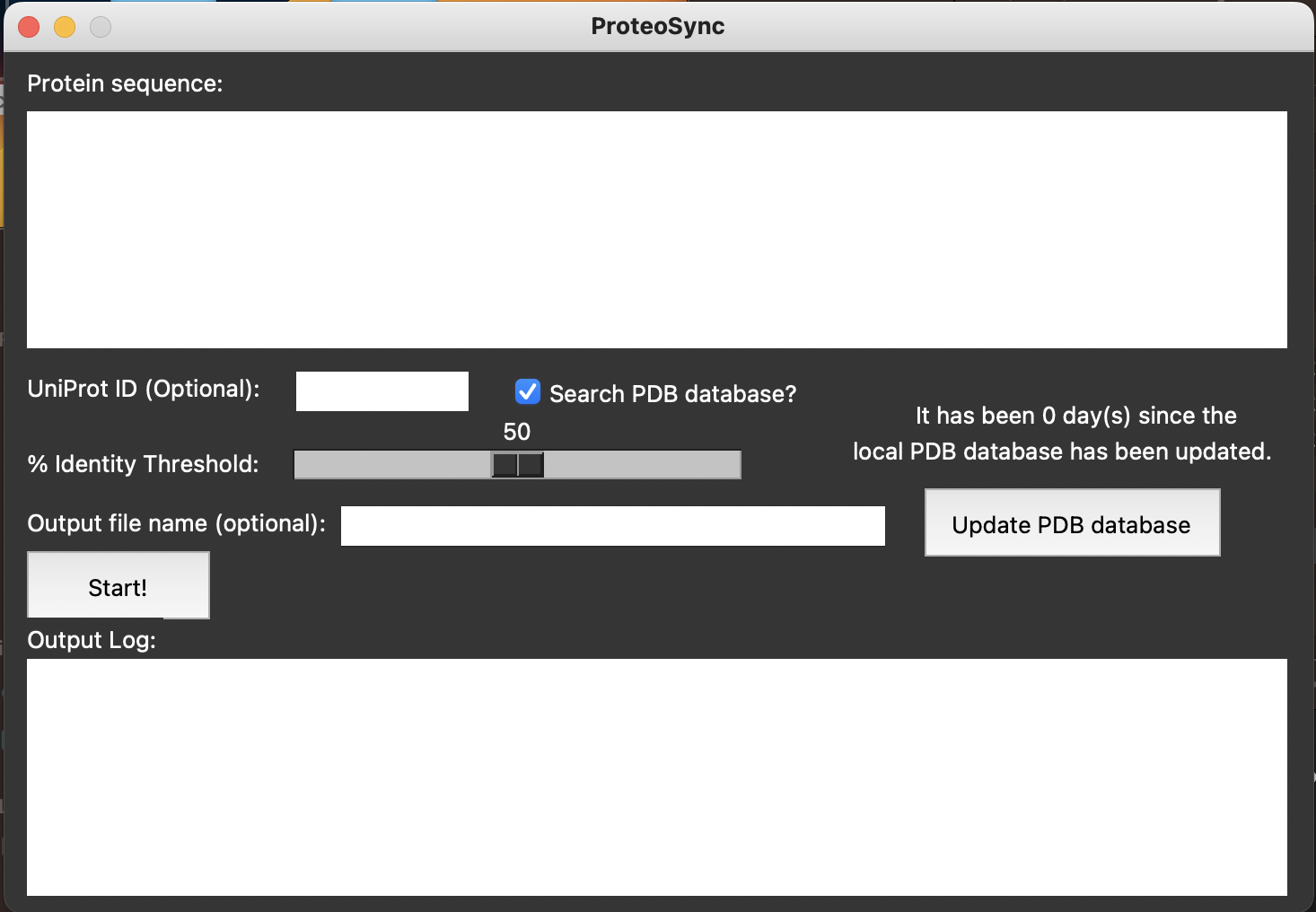
The program outputs a detailed sequence alignment, including clustalw conservation scores and the option to align sequences with both solved and predicted structures. It also generates a PyMOL script file that projects conservation scores onto a 3D model.
|
|
|
|
|
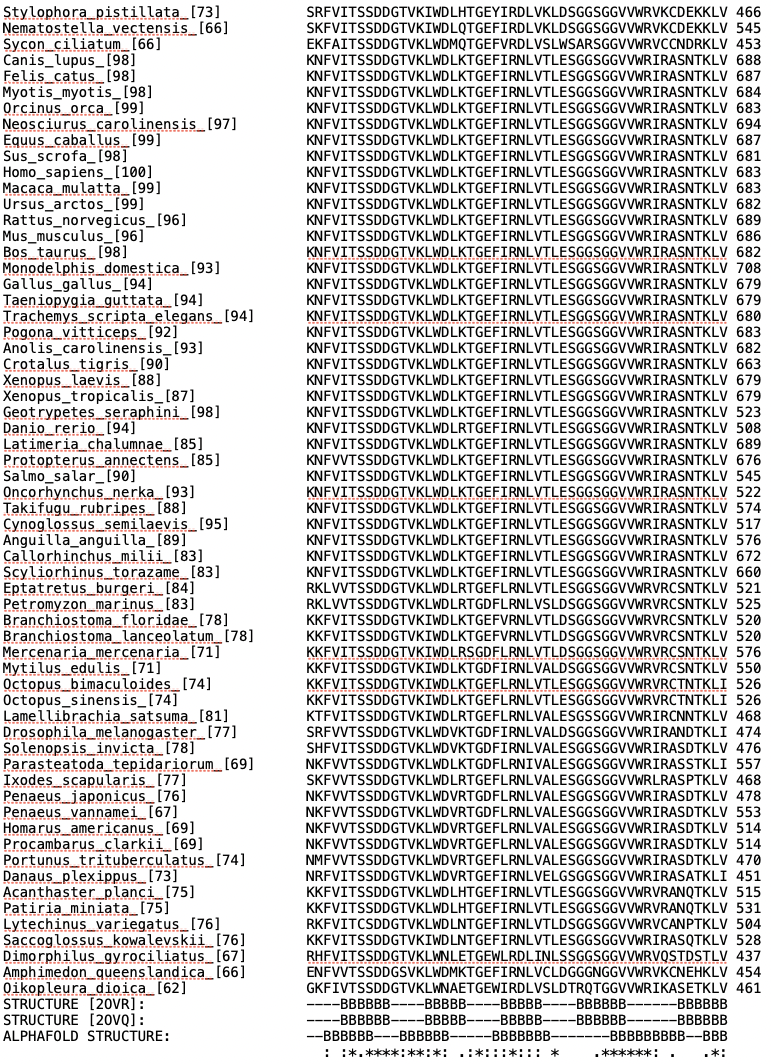
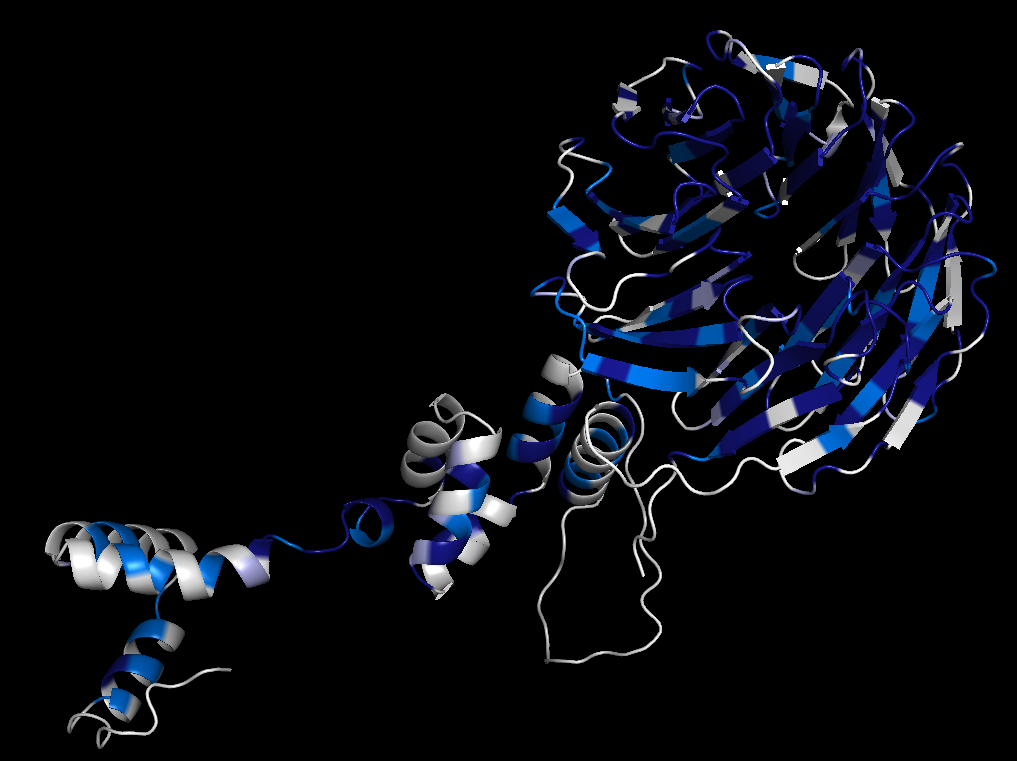
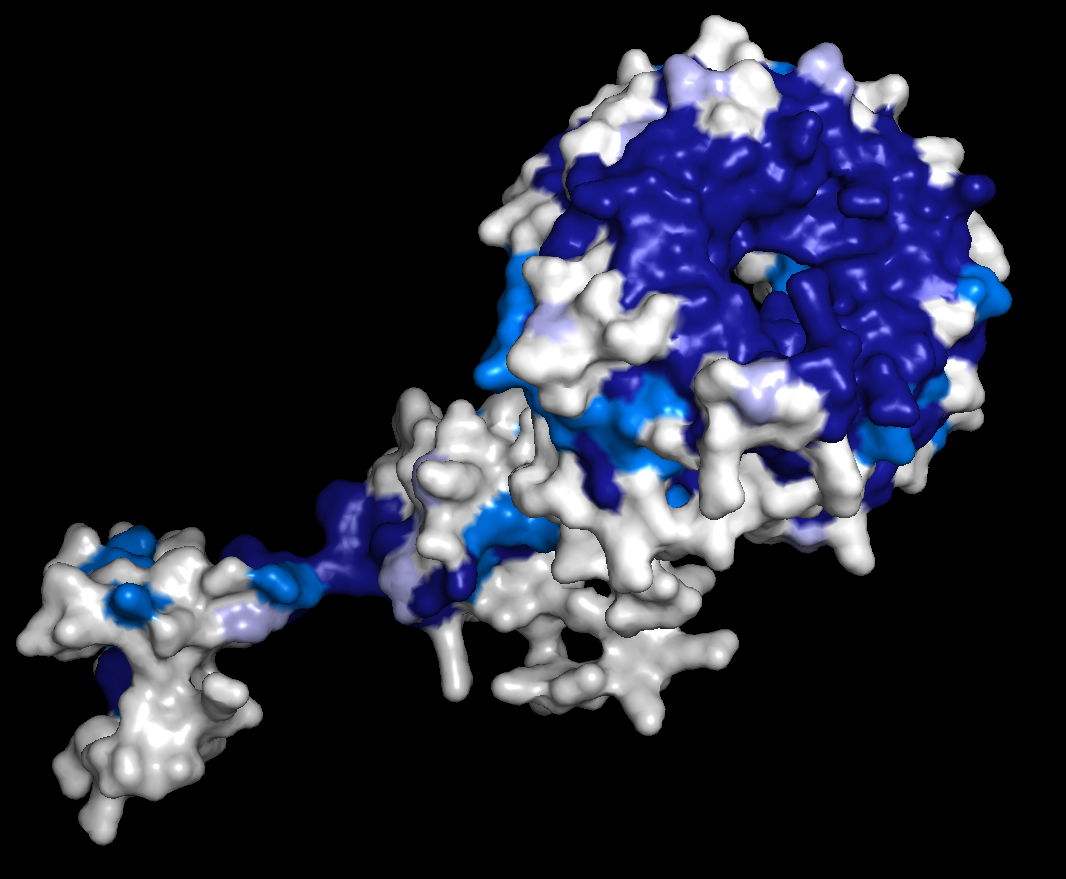
Left: Alignment output for the human tumour suppressor protein FBW7. Right: Conservation scores projected onto residues 222-707 of the AlphaFold preditcted structure of FBW7, with and without visible surfaces enabled. Darker colors correspond to higher conservation. A hot spot of conservation on the WD40 repeat domain of FBW7 identifies the functionally important degron binding pocket. |
|
For installation instructions, visit the GitHub page:
GitHub Link
Contact
For help, questions/inquiries and bug reports, please email elliot.sicheri@mail.utoronto.ca, I'll try to respond as soon as possible!